Automated Capillary Immunoassay System
Peggy Sue
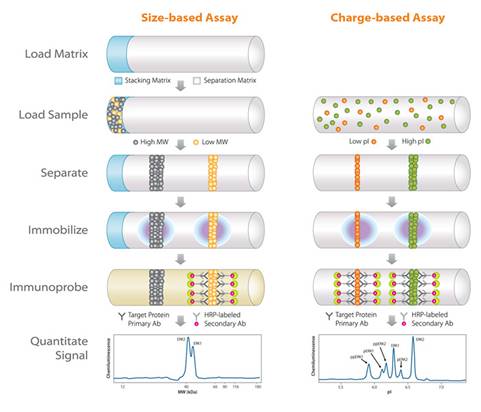
Two ways to get separation, identification and quantitation
Peggy Sue™ lets you separate and analyze proteins by size or charge from 2-440 kDa either by immunoassay or total protein. Got small sample volumes or starting materials? No problem. She uses as little as 0.2 µg/µL of protein in just 5 µL of sample, and runs up to 96 samples in one experiment.
A system for precise and accurate measurement of proteins and respective post-translational modifications in a format that is applicable to very small sample sizes..
Technology
• Employs high-resolution MW (size-based) or isoelectric-focusing (IEF, charge-based) separation, followed by target-specific immunoprobing to profile proteins and respective post-translational modification isoforms
• Fully automated and robust, provides bioanalytical labs with reproducibility and sensitivity in western blotting methods
• Have been applied for quantitative proteomic analysis in both discovery research and clinical practice
Assay Performance
• Precise and accurate measurement, digital data quantitation, good assay sensitivity, reproducibility and dynamic range
• Nanogram (ng) level protein loading, capillary platform allows quantitative proteomic analysis in extremely small and precious samples, such as stem cells, primary cells, fine needle aspirates, other patient specimens
• Increased sensitivity and specificity
• Multiplex analysis with fast assay turn-around time
• Straight forward assay standardization across multiple testing sites
Simple Western Assays
The assays provide custom pathway network profiling based on disease, drug target(s) etc. Assays identify critical pathways and molecular functioning mechanisms involved in the behavior of newly developed cell lines, PDXs, and clinical tumor samples.
The SpITR core has established over 200 targets identifying the following key wathways:
Apoptosis/Cell Death, Cell cycle and checkpoint control, Cellular metabolism, Chromatin Regulation/Epigenetics, DNA damage and repair, Gene regulation and DNA repair, JAK/STAT signaling, MAP Kinase signaling, NFκB signaling, PI3K/AKT/mTOR signaling, Protein Kinase C signaling, Receptor tyrosine Kinase signaling, Rho signaling, RNA regulation, TGF-β/SMAD signaling, Transcription regulation, Ubiquitin-proteasome, Wnt Signaling etc.
More Resources
Technology Overview: https://www.proteinsimple.com/simple_western_overview.html
High-throughput size-assay platform: https://www.proteinsimple.com/sally_sue.html#resources
Total protein assay for loading control: https://www.proteinsimple.com/documents/HowToGuide_Total_Protein_Normalization_RevC.pdf
Tested Samples with Simple Western Assays
Cultured mammalian cells:
breast, prostate, pancreatic, ovarian, liver cancer cell lines, and stem cells etc.
Tissue samples:
xenograft, mouse lung, mouse skin etc.
Clinical biopsies:
breast, prostate, pancreatic, ovarian, liver cancer cell lines, and stem cells etc.
Established MW-based Simple Western Assays
A panel of about 200 assays, covering key signaling pathways from receptor activation, down-stream signaling transduction, transcriptional regulation, cell cycle control to apoptosis etc., has been developed / established. This allows us to provide CCR investigators a signaling network array for protein activity characterization, therapeutic target identification and drug selectivity determination, as well as defining the regulatory mechanisms. We actively develop new assays based on the demands of the CCR/NCI/NIH community. Please contact us for more information.
Apoptosis/Cell Death:
Bad, phospho-Bad (pS112), Bak, Bax, Bcl-xL, BIM, Caspase 3 (full length, cleaved), Caspase 7 (full length, cleaved), Caspase 8 (full length, cleaved), cIAP1, cIAP2, FADD, FLIP, Mcl-1, PARP (full length, cleaved), RIP, SMAC/Diablo, Survivin, XIAP
Autophagy:
LC3A/B, SQSTM1/p62
Cell Adhesion and cell-matrix:
E-Cadherin, N-Cadherin (CDH2), EpCAM, Fibronectin, Integrin α5, Integrin αV, Integrin β1, Integrin β3, FAK, phospho-FAK (pY397), Caveolin-1
Cell cycle and checkpoint control:
Bmi1, EZh2, Rb, KID, PLK1, Cyclin B1, Cyclin D1, Cyclin D3, Cyclin E, E2F-1, MCM2, MCM4, MCM5, p27 Kip
Cellular metabolism:
AMPKα, phospho-AMPKα (pS485, pT172), PKM2, ACC, phospho-ACC (pS79), ATGL, phospho-ATGL (pS406), HSL, phospho-HSL (pS565, pS660), LPL, PPAR gamma
Chaperone proteins:
HSP70, HSP70α, HSP70β, HSP90
Chromatin and epigenetic regulation:
phospho-Histone H2A.X (pS139), DNMT1, STAG2, SMARCB1, TACC3, Histone H2A, HDAC 6, Acetyl-α-Tubulin (Lys40)
DNA damage/repair & gene regulation:
BLM, p53, phospho-p53 (pS15), phospho-Chk1 (pS345), DDB1, STAG2, Whip1, ATM, phospho-ATM (pS1981), PTIP, FANCD2
Epitope Tags:
GFP, GST, Flag
JAK/STAT signaling:
STAT3, phospho-STAT3 (pY705), STAT5, phosphso-STAT5 (pY694), JAK2
Loading Controls
α-Tubulin, GAPDH, β-Actin, Thioredoxin 1, ALAS1, HSP70, Vinculin, Glucose-6-phosphate dehydrogenase (G6PD), Rho-GDI
MAP Kinase signaling:
ERK1/2, phospho-ERK1/2, MEK1/2 and phospho-MEK (MEK1 pS217/221, MEK1 pT292, MEK1 pT386, MEK2 pT394, MEK1 pS298), p-90RSK 1/2/3, phospho-p90RSK (pT359), p38 alpha MAP Kinase, phospho-p38 MAP Kinase (pT180/182), JNK, phospho-JNK (pT183/185), JNK2, DUSP3, A-Raf, B-Raf, c-Raf,
NFκB signaling:
c-Rel, RelB, IκBα, phospho-IκBα (pS32/36), Iκκα, NFκB p65, phospho-NFκB p65 (pS468, pS536, pS529), NFκB1 p105/p50, NFκB2 p100/p52, TRAF1
PI3K/AKT/mTOR signaling:
pan AKT, AKT1/2/3, phospho-AKT (pS473), GSK3β, phospho GSK (pS9, pS21), PI3 Kinase, p110 α/β, PTEN, phospho-PTEN (pS380), mTOR, phospho-mTOR (pS2448), 4E-BP1, phospho-4E-BP1 (pT37/46, pT45, pS65), p70 S6 kinase, phospho-p70 S6 kinase (pT389), GSK3 alpha, Rictor
Protein Kinase C signaling:
PKCδ, phospho-PKCδ (pS299, pY311), PKCε, PKD1, phospho-PKD1 (pS744/748), PKCα, PKCβII, PKCθ, RasGRP3, phospho-RasGRP3 (pT133), PKD2
Receptor Tyrosine Kinase signaling:
EGFR, phospho-EGFR (pY1068, pY1173, pY845, pT992), HER2/ErbB2, phospho-HER2/ErbB2 (pY1248), HER3/ErbB3, GAB1, PLCγ1, Shc, phosho-Shc1 (pY239/240), Src, phospho-Src (pY527, pY416), phospho-tyrosine (4G10), FGFR-4, GRB10, IGIF, phospho-IGFIR (pY1135), VEGFR, MET, phospho-MET (pY1234/1235), PDGFRα
Rho signaling:
LIMK2, Cofilin, phospho-Cofilin (pS3), FHL1, FHL2, ROCK-1, ROCK-2, Rho-GDI
RNA regulation:
S6 Ribosomal protein, phospho-S6 Ribosomal protein (pS235/236, pS240/244)
TGF-β/SMAD pathway:
SMAD1, phospho-Smad1/5 (Ser463/465), SMAD2, phospho-Smad2 (Ser465/467), SMAD3, phopho-Smad3 (Ser423/425), SMAD5, SMAD4
Transporter, ion channel and vesicular trafficking proteins:
ABCG2, P-Glycoprotein (P-gp), CLIC4, Tsg101
Transcription regulation:
c-Myc (pS62, pT58), FoxO3A, phospho-FoxO3A (pS318/321), EGR1, cFos, Fra1, phospho-Fra1 (pS265), c-Jun, phospho-c-Jun (pS63, pS73), FosB, JunB, phospho-JunB (pT102/104), Jun D, CREB, phsopho-CREB (pS133), HMGA2
Ubiquitin-proteasome pathway:
Ubiquitin, Jab1, Ube2W
Wnt Signaling:
β-Catenin
